Darwin’s theory of natural selection was published in 1859, explaining how nature selects certain advantageous physical attributes (or phenotypes) to pass on to their offspring, so that they can better survive in their environment. Now over 150 years later, the theory is becoming a talking point in oncology, with natural selection within tumors helping to drive resistance mechanisms in cancer.
Like any natural selection process, evolution in cancer cells is known to create a large amount of diversity. This results in different patients with the same disease having genetically different tumors from each other. Also, within a single patient as cells divide and randomly mutate, evolution generates intra-tumor heterogeneity – with multiple genetically different cell lineages or sub-clones evolving in parallel within one tumor. Natural selection responds to environmental pressures, which in terms of cancer can be treatment such as chemotherapy. Cells which can survive treatment will multiply and pass on their traits to daughter cells, which can then cause resistance to the chemotherapeutic agent. Targeted therapies can also be stumped by cancer evolution – for example while a tumor may start as 99% wild type for a protein such as KRAS or EGFR which confers sensitivity to a specific agent, 1% of the tumor proteins may harbor a mutation which confers resistance. As treatment continues, the mutated resistant cells will take over the tumor rendering the treatment useless.
So, what are the treatment options if cancer can essentially always evolve its way around a treatment? Using a drug cocktail (similar to HIV treatment) might be a good start. Instead of using agents sequentially and changing treatment when resistance occurs, agents that target different genes can be used together from the start of treatment. It takes longer for resistance to emerge if multiple mutations to work around multiple therapies are needed. New therapies can also be designed that could potentially “direct” natural selection of cancer cells to prevent the outgrowth of resistant cancer populations. Research is also ongoing to develop new highly sensitive methods to detect which sub-clones are within each tumor, which could be used to predict which treatment a patient will respond to, and how evolution of resistance might occur.
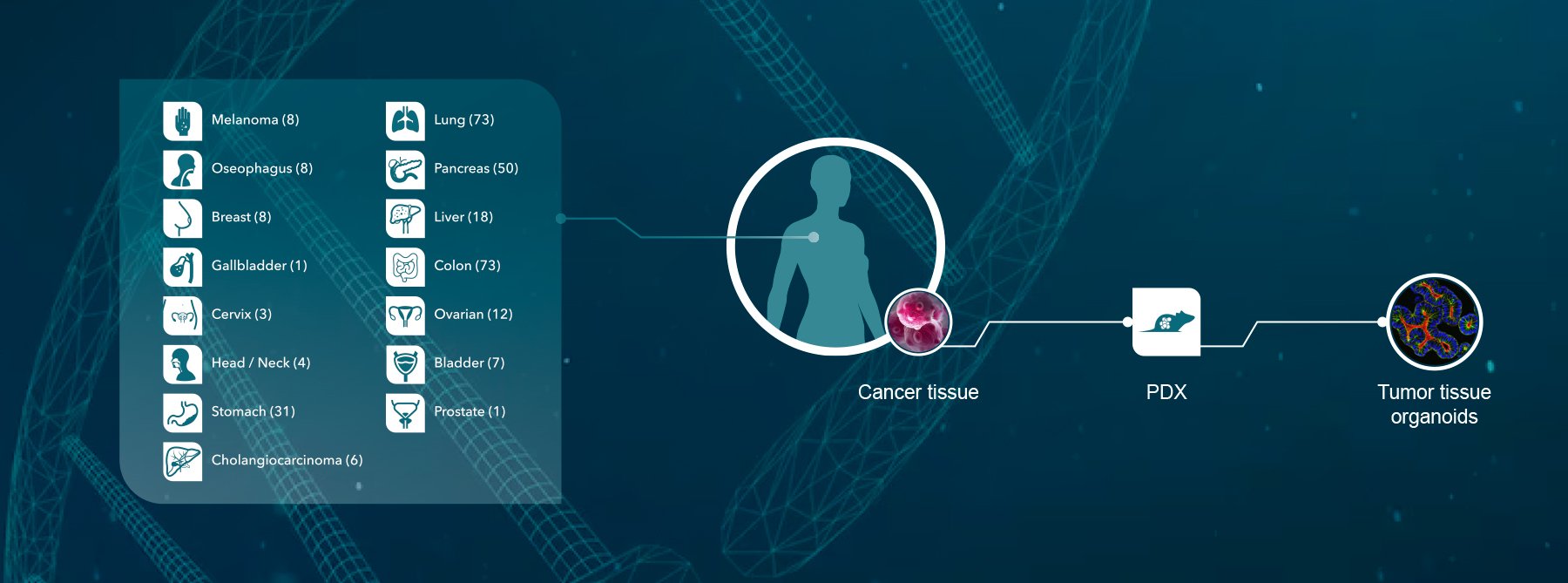
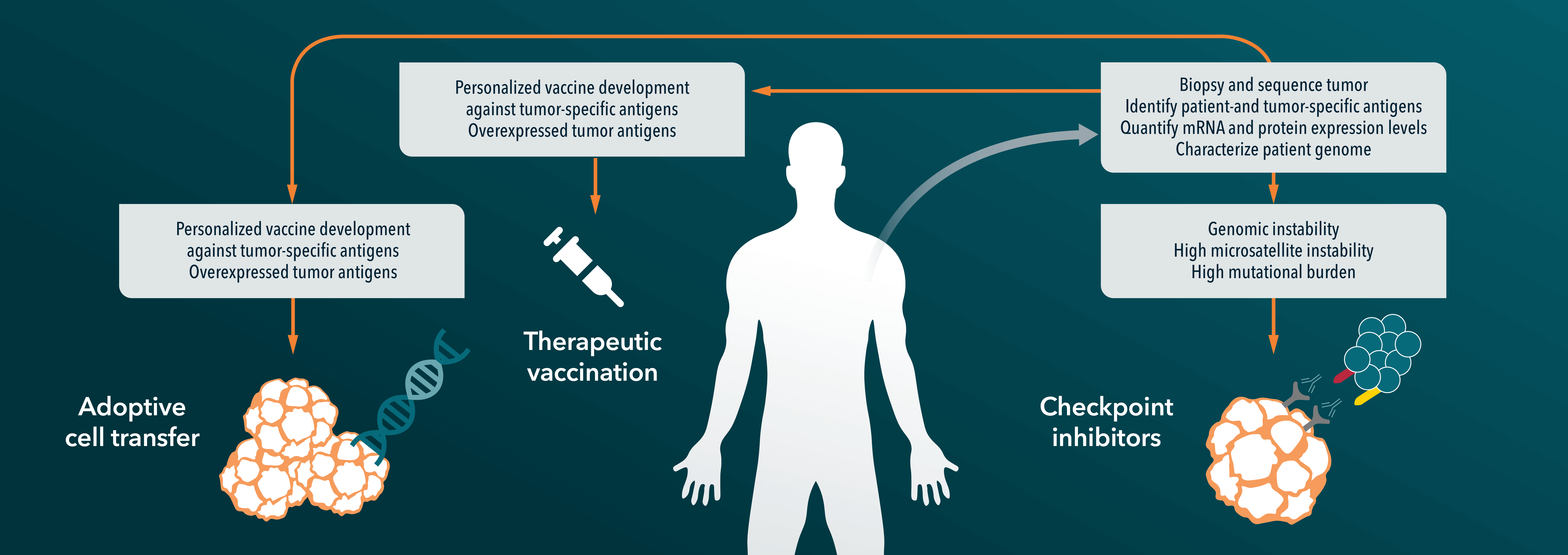
Personalized medicine has long been hailed as the future of oncology, and will become even more important in cancer evolution treatment paradigms. Cancer genomics could hold a lot of answers and as DNA sequencing reaches the magic $1,000 a genome cost, could become a more routine predictive tool. Efficient translational tools will also be required to model resistance mechanisms and to identify combination treatments which could be carried through in to the clinic. Patient-derived xenograft models (PDX models) are an ideal candidate for this. They preserve the biological characteristics and genetic diversity of the original tumor, so can be treated with standard of care and targeted agents to see how resistance develops, as well as being used to trial new agents and combination regimens to target resistance. Combining a large number of patient-derived xenograft models in a Phase-II like clinical trial can also be used to identify groups of patient responders within an indication, as well as biomarkers of response, to guide which patients would be most likely to respond to any new agent or combination.
Crown Bioscience have pioneered the use of patient-derived xenograft models, and now have the largest commercial collection of around 1,300 well-characterized models (HuPrime®), which are currently also undergoing RNA sequencing. Within this collection we also have a range of models resistant to radiotherapy, chemotherapeutics, and targeted agents, which can be used to study novel treatment regimens and to elucidate mechanisms of action of resistance. Contact us at busdev@crownbio.com to talk to our experts on how we can further your patient-derived xenograft model research and your translational oncology needs.