Dr Davy Ouyang, CrownBio Vice President of Scientific Research & Innovation answers the most popular questions from his recent syngeneic model webinar, covering the latest research, model developments, and platforms for moving past syngeneics for advanced immuno-oncology studies.
Dr Davy Ouyang, CrownBio Vice President of Scientific Research & Innovation answers the most popular questions from his recent syngeneic model webinar, covering the latest research, model developments, and platforms for moving past syngeneics for advanced immuno-oncology studies.
Syngeneic Study Design and Immunoprofiling
Why did you choose a 100-500mm3 tumor volume for syngeneic immunoprofiling?
We have performed comprehensive longitudinal immunoprofiling of syngeneic tumors at different volumes to cover larger dosing windows. Our initial profiling presented here used approximately 100mm3 tumors as that is the tumor volume when our treatment starts.
Can you perform cytokine analysis in these models?
We have run a lot of cytokine profiling across various syngeneic models as pharmacodynamic readouts within our client studies. We provide both MSD and Luminex assays for these analyses.
What time point do you use for proteomic and RNAseq analyses?
Our endpoint presented here was our efficacy endpoint, 21-28 days into treatment.
Using isotype controls – do you see any efficacy changes in your syngeneic models?
We don't see any isotype control effects in our syngeneic studies. We have completed a comprehensive study with 12 syngeneic models receiving anti-PD-1 treatment, where we setup both PBS and isotype controls for each model. The growth curves of the 2 control groups always merge together.
Do you have a syngeneic model relevant to translate findings for cold tumors like B16-F10?
We routinely use the Lewis lung model LL/2 for this.
Which of the discussed models do you think is the most translational?
I think tumor homografts (discussed in more detail below) have more translational value in terms of mimicking human tumor morphology, TME, and driver mutations.
Immune Cell Depletion of Syngeneic Models
For which models do you have immune profiles for TILs following depletion?
We have data on 4 syngeneic models: MC38, Hepa 1-6, CT26.WT, and EMT6.
Specifically looking at FACS analysis of TIL following immune cell depletion in the MC38 model. CD8 T cells are increased after CD4 depletion. Therefore, I presume the stable frequency of CD3 T cells after CD4 depletion is due to the CD8-T increase. But, why do the CD3 T cells look stable when CD8 T cells are depleted, although the CD4 T cells are not changed after the depletion?
After CD8 depletion, there is a subtle increase in CD4 T cells, and slight drop of CD3 T cells.
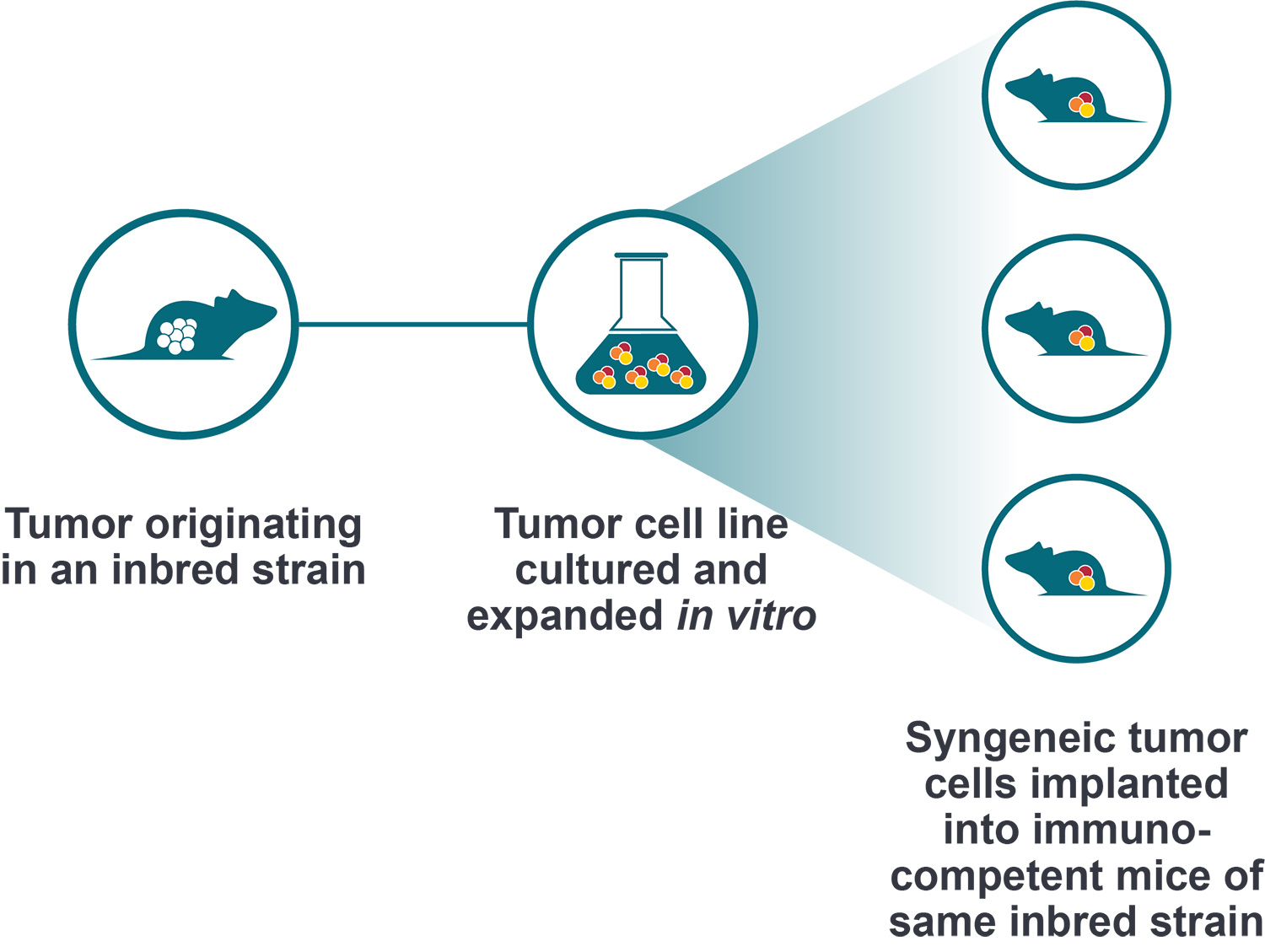
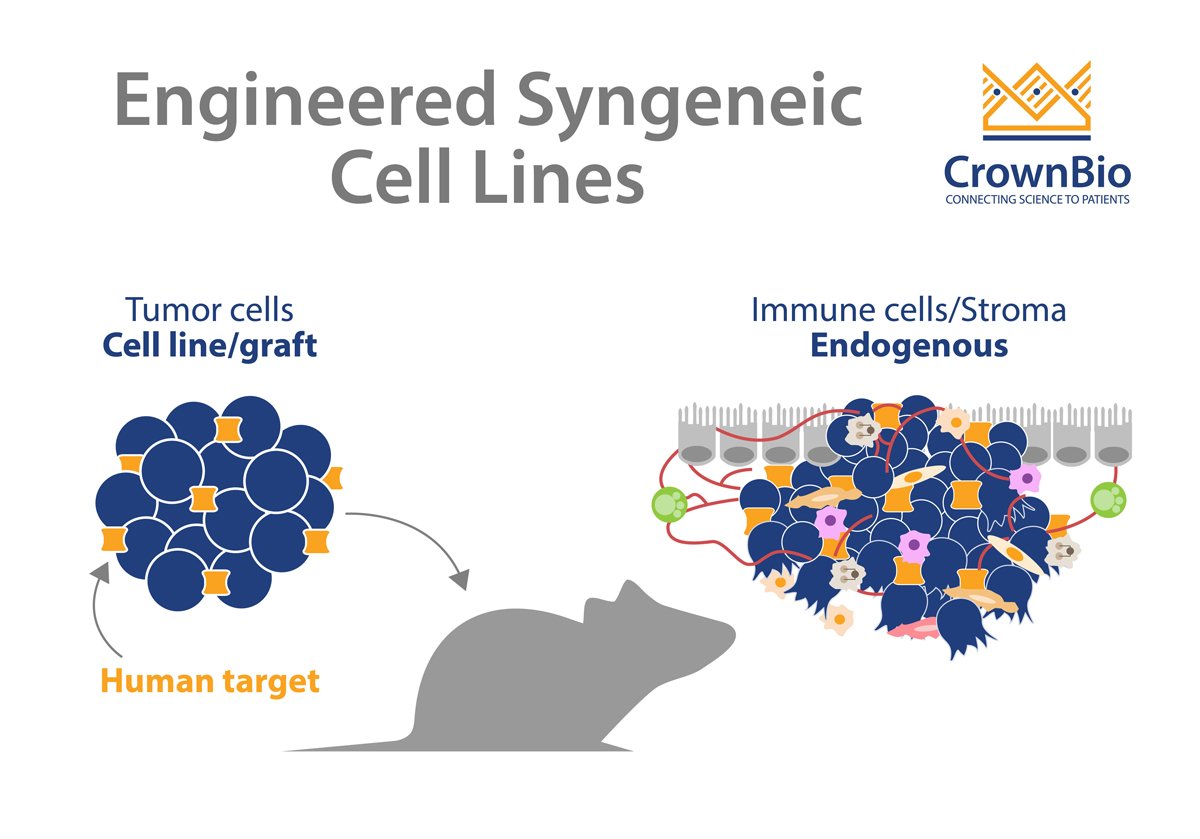
Engineered Syngeneic Cell Lines
You presented differences between MC38 vs MC38-OVA and some nice characterization data. Do you have the same approach for cell lines with and without luciferase?
We are in the process of profiling all our of luciferase-tagged cell lines. Data so far indicates that the introduction of any antigens e.g. OVA or hTAA, all lead to increased CD45 and T cells in the TIL. I think that the introduction of luciferase to a cell line will induce the same result.
Do you have tumor-antigen engineered syngeneic lines where the T cell response is much more suppressed than in the MC38 line? Maybe CT26.WT that has a more Th2/M2 macrophage environment?
So far, all hTAA over-expressing lines seem to have increased immunogenicity and better T cell responses. However, we have generated some PD-1 resistant lines such as B2M KO, STK11 KO, JAK1/2 knockout.
Tumor Homograft MuPrime™ Models
How are the tumor homograft models generated?
This is a similar operation to generating patient-derived xenografts (PDX). A number of early passage tumor chunks are banked for propagation. We then set up studies with seed tumors.
How much immunoprofiling data do you have for your tumor homograft models?
We have immunoprofiling data for around 40 tumor homograft models now, which is always increasing.
Do you see a difference in TIL profile between subcutaneously and orthotopically implanted KPC models (or other tumor homograft models)?
Yes, there are some difference in MDSC and macrophage infiltration. Other features can be similar -- for example, lack of CD8 T cells, relatively high B cells, etc.
The KPC model immunoprofiling looks like it has essentially no CD8+ or Tregs. Is this correct?
There are almost no CD8 T cells in this model.