Are you confident that your cell lines and biological models are really what you think they are? Do you know that common PCR-based cell line authentication services may not be able to detect contamination of up to 20% of your cell line?
In this post, we highlight the value of using next-generation sequencing (NGS) as a higher sensitive, accurate and throughput, and multifunctional approach for cell line authentication and contamination detection that has been shown to outperform conventional PCR-based STR assays.
Why Does Cell Line Authentication Matter?
Misidentification and contamination of biological models—including cell lines, organoids, and homograft and xenograft models—remains a challenge. Studies indicate that up to 33% of popular cell lines are contaminated by intra-/interspecies cells, mycoplasma, and/or viruses, and the International Cell Line Authentication Committee (ICLAC) currently lists 530+ misidentified cell lines (as of June 2021) that have no known authentic stock.
The use of misidentified cell lines continues to be a big problem, with Horbach & Halffman (2017) reporting that more than 32,500 papers referred to data from cell lines the ICLAC had classified as misidentified.
Contamination also continues to be pervasive, and it can arise from a variety of sources including cross-contamination with other cell lines and microorganisms (e.g., mycoplasma and/or viruses).
Reproducibility is one of the hallmarks of the scientific method and using misidentified and/or contaminated cell lines can lead to unreliable data (Fig.1). Thus, skipping the authentication step can result in inaccurate conclusions, retraction of publications, and wasted time and money if the cross-contamination or misidentification is found to invalidate the data.
The financial cost of invalid research caused by misidentified cell lines has been estimated to exceed $50 billion. It is therefore not surprising that certain funding agencies such as the NIH and more publishers are requiring researchers to provide biosample authentication.
The U.S. FDA also requires that all materials included in an investigational new drug (IND) application are validated.
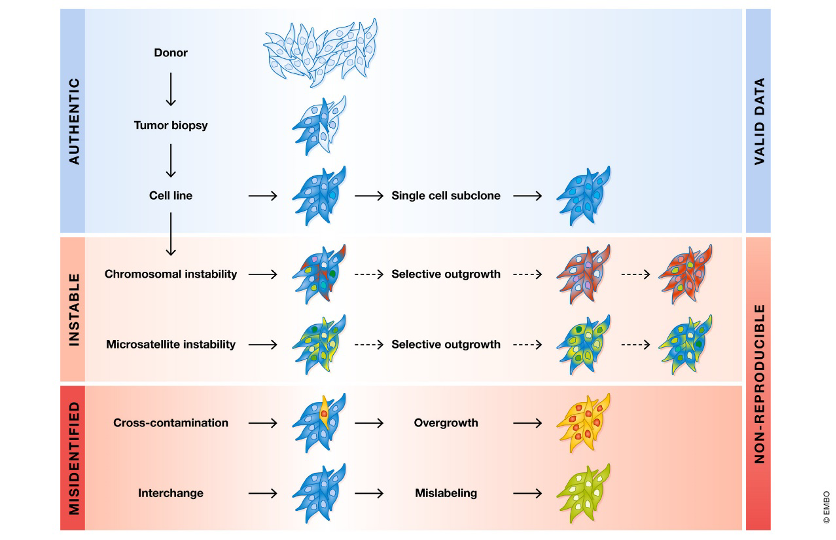
Fig 1. Scenarios of data generation using cell lines as tumor models (The EMBO Journal (2022)41:e111307)
Here are the major reasons why researchers should consider authenticating their biosamples:
- When a new biosample is acquired or developed
- As a standard quality-control measure
- When in doubt
Conventional Multiplex PCR-based STR Assays for Cell Line Authentication and Their Limitations
PCR-based STR assays use primers designed to recognize repeated DNA segments of 2-6 base pairs in length. Since the targeted segments vary within a given population, a DNA ‘fingerprint’ can be generated. This technology has been extremely valuable for authenticating cell lines and tracking the identity of human tumors samples that are either derived from patient xenografts or cell lines.
Depending on the vendor, STR assays generally target only 9 to 24 loci, and limitations with PCR technology also restricts the sensitivity and accuracy of the cell line authentication. The accuracy of an STR assay can be especially low in cases where there is a close genetic relationship, such as when trying to authenticate mouse cell lines derived from specific strains of inbred animals that lack unique genetic markers or different tumor cell lineages from the same human donor.
Additionally, cell lines with mutations in mismatch repair (MMR) genes are known to exhibit microsatellite instability and a hypermutator phenotype. Consequently, this can lead to genetic drift and/or outgrowth of contaminating cells and STR misclassification.
The sensitivity of PCR-based cell line authentication assays are claimed to be in the range of 5-10%, though this is variable and depends on a variety of factors including the nature of the cell lines being tested and the quality of the data, however, in reality, sometimes the PCR-based CLA assays can not pick up contaminations up to 20%.
Cell Line Authentication with Deep Sequencing
Significant technological advances have resulted in substantial reductions in the cost of genome sequencing. This includes NGS-based technologies which are amenable to high throughput and multifunctional analyses, including for authenticating large biobanks of cell lines which are notorious for harboring lines that are cross-contaminated.
It is now well-established that NGS significantly outperforms traditional profiling methods for characterizing mouse and human samples, including cell lines, organoids, xenografts, and patient tissues.
At Crown Bioscience, we use deep next-generation sequencing (NGS) with a panel that encompasses 600+ SNPs and chromosome segments to accurately characterize mouse and human samples, at 3000x coverage.
In addition, by using advanced bioinformatics and statistical modeling to support NGS-based cell line authentication, both the identify and frequency of the SNPs can be obtained. And since hundreds of samples can be profiled in a single run, this represents a major improvement over conventional STR assays, which are low throughput.
In terms of coverage, high throughput capability, and sensitivity, NGS is superior to the PCR-based STR assays for cell line authentication which only targets 9 to 24 loci.
Another advantage of NGS lies in its multifunctional capabilities that are not possible with PCR-based STR assays. For instance, this NGS method can also be used to:
- Detect viruses and mycoplasma.
- Determine human gender, and other human genetic elements.
- Trace genetic drift and reconstruct sample phylogenies.
- Determining the contamination ratio of mixed samples.
We have generated a SNP fingerprint library for the most commonly used oncology research models, which cover more than 1,200 human cancer and 30 mouse cell lines.
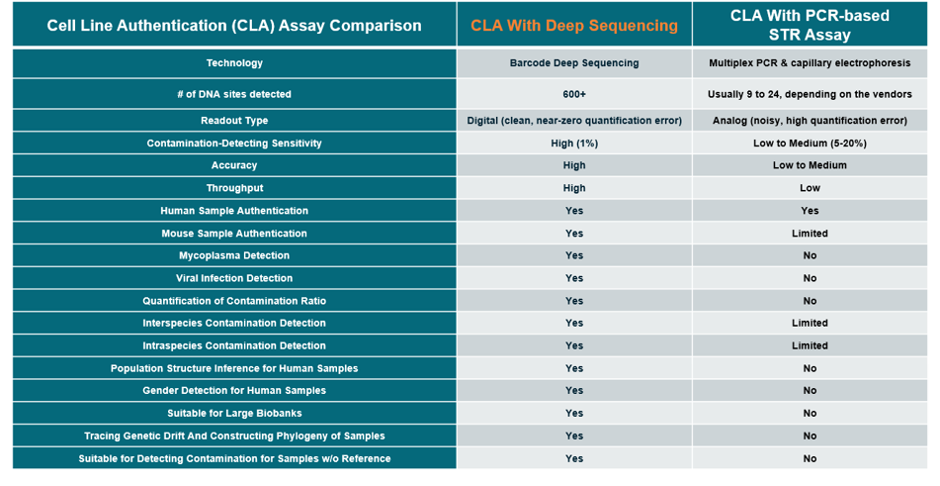
The following table compares the deep sequencing and PCR-based STR assays for cell line authentication:
Conclusion
Cell line authentication is increasing in importance funding and regulatory agencies, journals, and the scientific community are requiring researchers to demonstrate the authenticity of their biological samples.
NGS-based SNP profiling is a high throughput, multifunctional, and cost-effective method for cell line authentication. Our unique NGS-based biosample authentication panel encompasses over 600+ SNPs and other chromosome segments that can accurately characterize mouse and human samples including cell lines, organoids, xenograft models, and patient tissues. We have also generated unique DNA fingerprints for the most commonly used oncology research models, covering over 1,200 human cancer and 30 syngeneic cell lines.
If you are interested in learning more about how to use NGS-based profiling for your biosample authentication needs, please visit our Cell Line Authentication page, or contact us by email now.