Applications and Benefits of Engineered Syngeneic Cell Lines
 Explore the use and main benefits of engineered syngeneic cell lines for investigating drug targets and biological mechanisms of action.
Explore the use and main benefits of engineered syngeneic cell lines for investigating drug targets and biological mechanisms of action.
Using Syngeneic Tumor Models for Immunotherapy Studies
Syngeneic tumor models, with an intact and functional immune system, are the most common immunocompetent models in preclinical immuno-oncology studies. Generated by inoculating mouse cancer cell lines into a genetically identical murine host, syngeneics effectively capture the dynamic interplay between the immune system and tumors.
Syngeneic models also respond to clinically relevant checkpoint inhibitors, both as monotherapies and combination regimens. The pharmacology of currently used checkpoint inhibitors was first tested in syngeneic tumor cell-line-derived models. For example, the clinically relevant combination of anti-CTLA-4 and PD-L1 checkpoint inhibitors, were shown to have enhanced antitumor activities when used in combination in several mouse syngeneic colorectal tumor models.
Overall, syngeneic tumor models possess several key attributes that make them highly amenable for immunotherapy studies:
- They have intact and competent immune systems.
- They rely on syngeneic tumor cell lines that can be rapidly and reproducibly expanded in vitro prior to implanting into wild-type hosts.
- They are scalable and can be efficiently used in large-scale studies.
- Many of them have robust historical growth and drug sensitivity data, which is important when it comes to properly interpreting study data.
- Syngeneic cell lines can be engineered to express specific factors, such as a target antigen.
In this post, I’ll focus on the ability to engineer syngeneic cell lines, and advantages these models can bring for preclinical immunotherapy studies.
Engineered Syngeneic Tumor Cell Lines
Engineered syngeneics are modified to express specific genes or modulate gene function. This provides cell lines for in vitro studies, for looking at the biological mechanisms of DNA repair, cell cycles, and antigen expression. Engineered syngeneic models are also valuable for validating target-specific therapeutic agents in vivo.
Engineered expression of specific targets also enable a better understanding of how specific targets affect drug response or resistance, such as by making tumors “hot” or “cold" - hot tumors are inflamed and elicit a response from the immune system, whereas cold tumors do not. Ultimately, this insight into the underlying mechanisms in drug responsiveness helps with target identification and validation. That, in turn, can lead to better biomarker and treatment strategies, where patients can be stratified based on biomarkers of drug response.
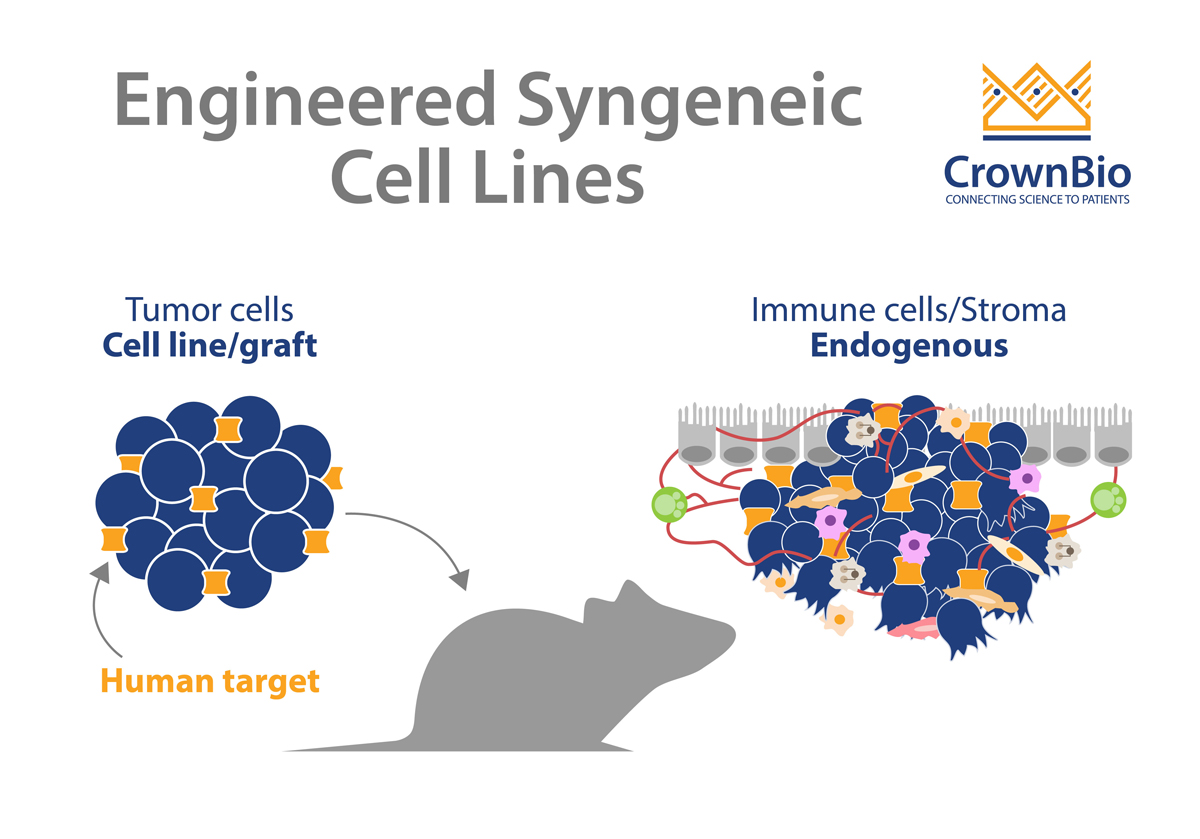
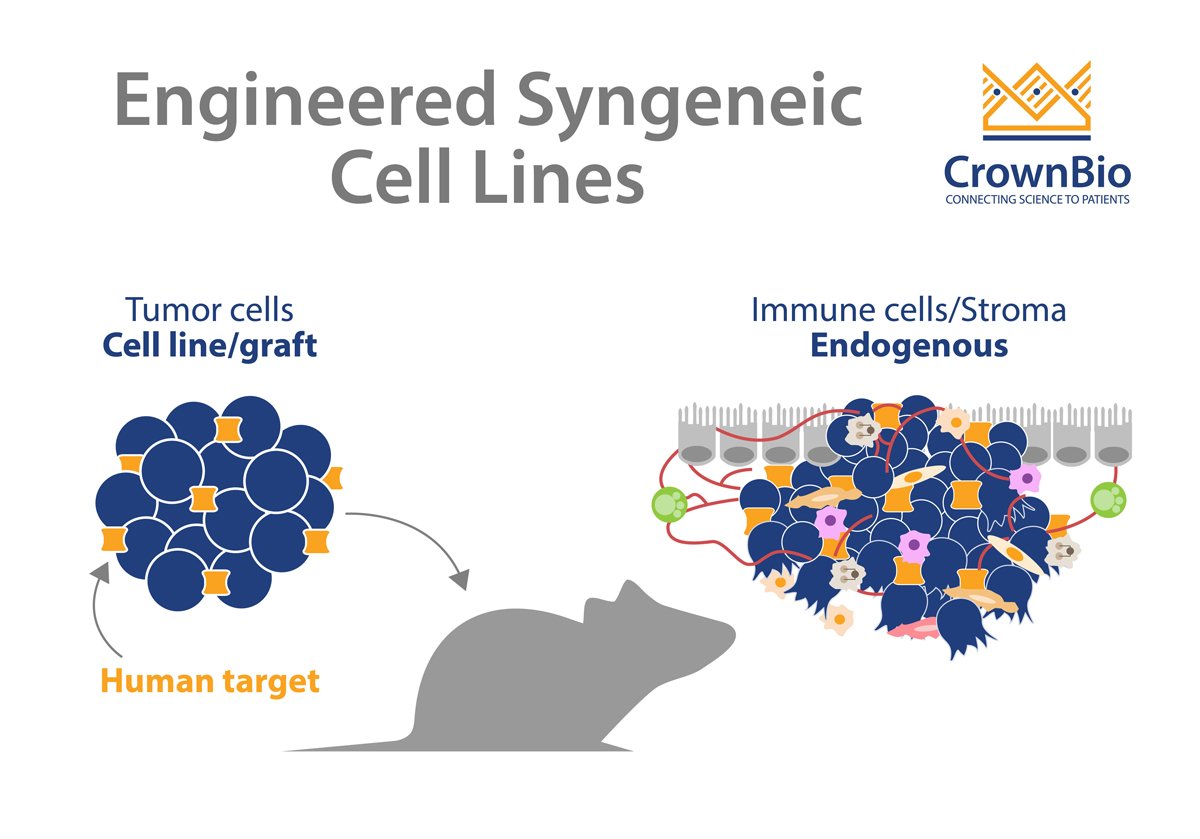
Classical syngeneic models show only murine target interactions. With the help of engineering, these models are instead optimized to genetically humanize a target of interest in the tumor cell lines. For example, catalogs of mouse cell lines have been engineered to express human targets such as hPD-L1, HER2, EGFR etc. These engineered cell lines can then be assessed for in vivo growth curve and baseline immune profiled.
Role of Engineered Syngeneic Tumor Cell Lines in Immunotherapy Preclinical Studies
Engineered syngeneic tumor cell lines expressing tumor antigens or checkpoint inhibitors have been instrumental in immunotherapy preclinical studies. For example, immune checkpoint proteins, such as PD1, PD-L1, VISTA, Tim3, and Lag3, are all known to prevent tumor killing by T cells. Therefore, preclinical immunotherapy studies have been designed using engineered syngeneic tumor cell lines expressing these immune checkpoints, to screen high affinity antibody reactivity against the expressed protein.
Since not all models effectively respond to immune checkpoint inhibitors (which may be due to their intrinsically low immunogenicity), tumor cells can also be engineered to express highly immunogenic proteins, such as ovalbumin (OVA). Such an approach renders the cells more immunogenic and is an efficient way to generate additional models for immunotherapy research to investigate whether functional enhancement of T cells can improve the antitumor reactivity of tumor specific T cells.
Conclusion
Since they have an intact and competent immune system, syngeneic tumor models have been extremely beneficial for preclinical immuno-oncology studies. Engineering syngeneic cell lines provides an expanded and humanized range of targets, to better investigate drug targets and explore biological mechanisms of action.
Cite this Article
Pal, R., (2020) Applications and Benefits of Engineered Syngeneic Cell Lines - Crown Bioscience. https://blog.crownbio.com/applications-and-benefits-of-engineered-syngeneic-cell-lines