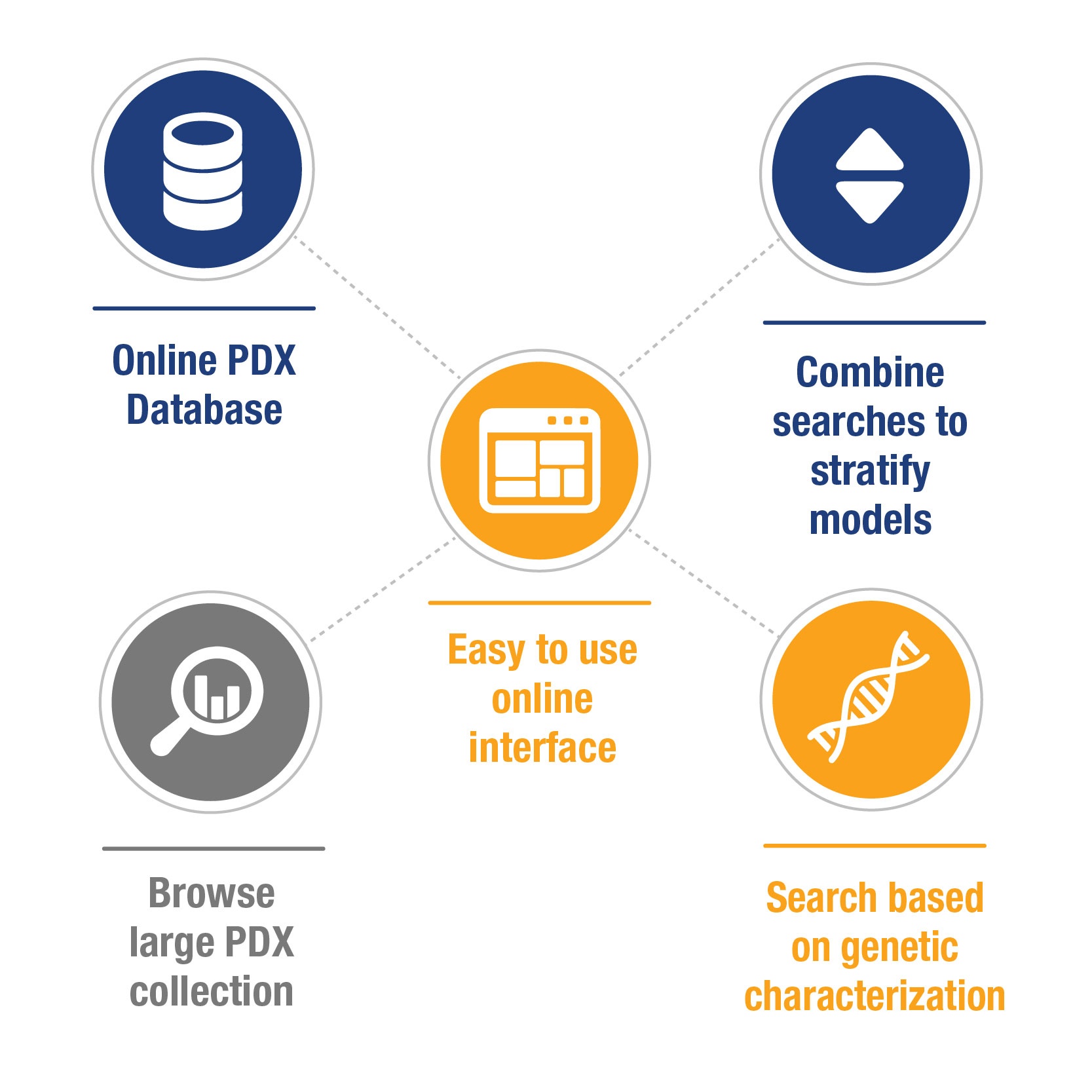
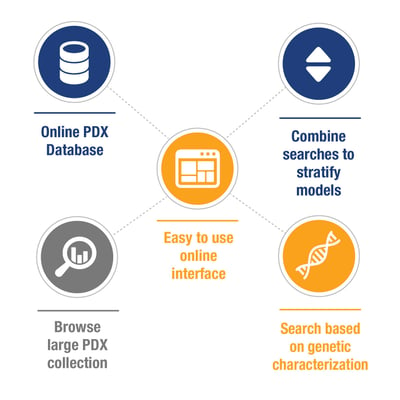
 Starting to write this blog, I realized it could also be titled ‘What to Expect From a Good PDX Database’, as the easiest way to find and choose a model to match your study needs is to search through an online collated database of PDX, which allows for a wide variety of model parameters to be examined.
Starting to write this blog, I realized it could also be titled ‘What to Expect From a Good PDX Database’, as the easiest way to find and choose a model to match your study needs is to search through an online collated database of PDX, which allows for a wide variety of model parameters to be examined.
Essentially, how you choose depends on what you are looking for – a particular cancer subtype, a fusion your inhibitor targets, or maybe drug resistant vs sensitive models. Having a large collection of PDX, collated via background information and searchable through characterization data should make it easy to stratify from a couple of thousand models down to exactly what you need.
Simple Browsing, Searching, and Model Stratification Required
There are many factors which help decide the models you need, both in patient background information and for developed models. Some people want to browse a full collection of breast cancer PDX to choose a panel, others want to directly look at all models treated with cisplatin, for example.
To answer these needs, patient background details and simple model information (pathology, IHC, growth data, treatment data) need collated in a user friendly manner, allowing both browsing and easy stratification. Grouping models by cancer type allows database users to see similar model information side by side, and easily select by browsing factors such as:
- Patient ethnicity
- Treatment naïve vs pretreated patients
- Disease subtype including simple IHC protein expression data
- Growth rates
Simple searching options can include selecting models (by cancer type or across all cancer types) by:
- Treated vs untreated models
- A specific drug treatment
- Models with profiling data available for more in depth searching
Searching by Characterization Data
Some researchers have specific genetic needs when looking for models, and these should also be accommodated within a PDX database. Historically, characterization data would have been generated by microarray for gene expression and copy number, now this is more likely to be covered by RNAseq and WES, respectively. Databases usually allow searching for:
- Gene expression
- Gene copy number
- Gene mutation
- Gene fusion
- miRNA expression
This allows users to select models directly based on the characteristics they need e.g. models across all leukemia types with BCR-ABL fusions, breast and ovarian cancer models with BRCA mutations, or combinations of searches such as lung cancer models with EGFR mutation but normal expression levels, or top 10% of models expressing gene A which are also the bottom 10% of models expressing Gene B. Searches like these need a large proportion of a PDX collection to have been sequenced, but can provide a rapid method to stratify complex needs and find models to fully mimic the clinical situation.
Selecting Models from Tumor Microarrays
Another way to search for models of interest is via TMAs. They allow rapid and high-throughput assay screening and identification of molecular targets, diagnostic, and prognostic markers of interest. This can be at the DNA, mRNA, or protein level, under identical, standardized conditions.
This screening method can be integrated with a PDX database – with models included on premade TMAs highlighted, allowing panels of models to be preselected and then subsequently screened, usually across one cancer type per TMA.
Choosing the right PDX model to match complex study needs may initially seem difficult, hopefully, this blog has shown with the help of a good database it should be a quick and easy process.