 Explore the emerging field of immunogenomics and how it’s being used to answer key questions related to immunotherapy response and resistance.
Explore the emerging field of immunogenomics and how it’s being used to answer key questions related to immunotherapy response and resistance.
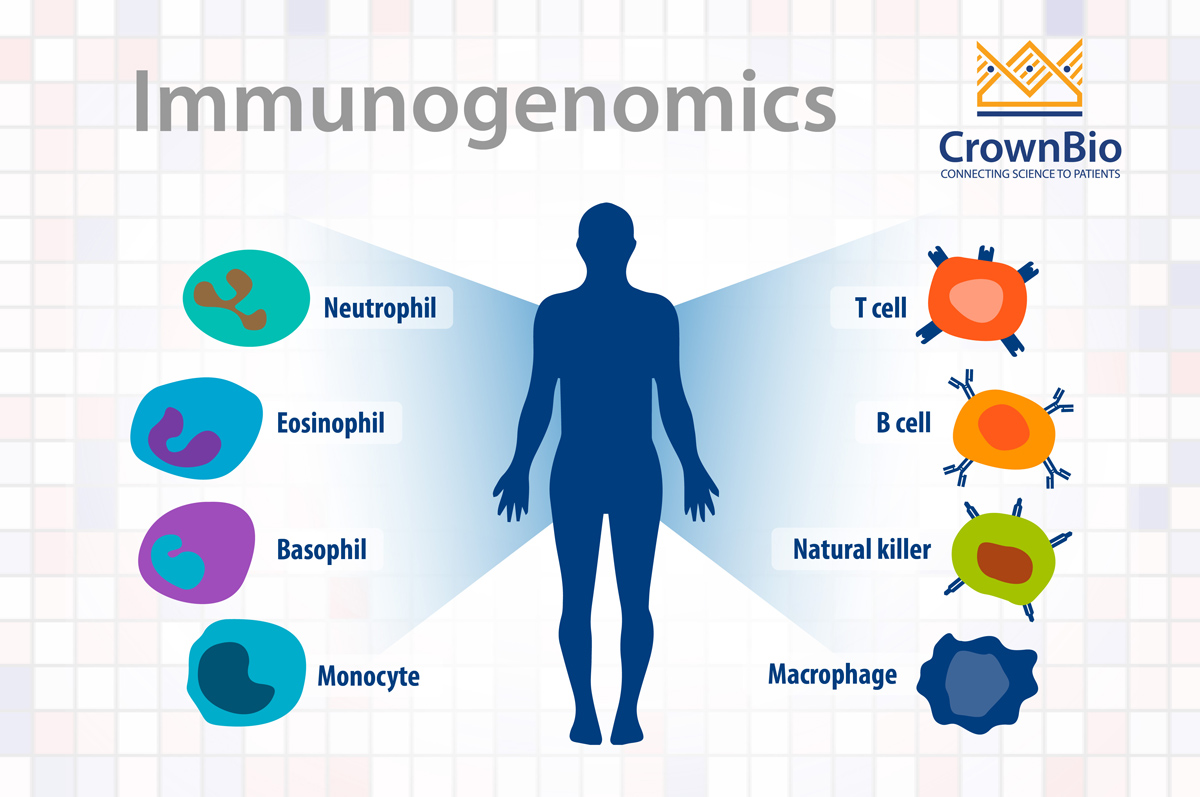
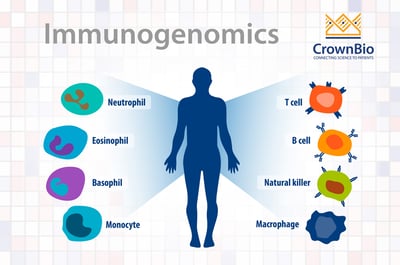
What is Immunogenomics?
The genomics of tumor cells have been the focus of cancer research and drug development for many years, based on their germline alterations and somatic mutations. This has resulted in the molecular changes in the surrounding tumor-associated immune cells not being investigated in depth.
Immunogenomics is a new field, combining the parallel study of both tumor cell and immune cell genomics to establish biomarkers to bridge the gap in response and non-response to immunotherapies.
The Principles of Immunogenomics
Cancer cells are generated, and become immortal, due to healthy cells losing their ability to control growth. The process can be a result of inherited DNA mutation or changes in DNA mutation over time.
The immune system provides protection and surveillance from cancer and other infections. But, that same immune system can also facilitate the growth of tumor cells, known as either equilibrium, or escape in the case of the immune system not recognizing the tumor as foreign.
Immunotherapies have emerged as leading treatments to reinvigorate the immune system from escape or to restore equilibrium, resulting in inhibition of, or slowing, tumor growth. Immunogenomics helps us uncover functional changes in the immune microenvironment caused by immunotherapy resulting in patient drug response or non-response.
Immunogenomics allows improved understanding in changes to the immune environment by looking at expression profiles of immune related genes before and after immunotherapy treatment. In parallel, immunogenomics can provide the mutations, as well as neoantigens, in a tumor which change over the course of immunotherapy treatment. All of this information helps us to understand why the tumor escapes or maintains growth and whether we can customize immunotherapies for individual patients.
The Benefits of Immunogenomics
Using immunogenomics allows us to investigate more about the immune response to cancer. Questions we can hope to answer using immunogenomics include:
- The role of immune cells which might negate the effects of therapies on tumor cells
- Why some immunotherapies work effectively against tumors and why some drastically fail
Information gained from the immunogenomics of immune cells also helps to tailor immunotherapy as a personalized medicine, based on specific patient requirements. This will allow patients to experience the maximum possible benefits from immunotherapy with fewer side effects.
Available Immunogenomic Technologies
Various sequencing methods are used to characterize the phenotype for both tumor and microenvironment interactions with an emphasis on immune cells. NGS-based methods such as RNAseq and WES are commonly used due to their high throughput capabilities and decreasing cost.
These platforms provide a depth of information suited for both exploratory and hypothesis-based approaches. Gene panels based on NGS and other technologies such as NanoString are useful for a more targeted approach. Newer technologies enable the coupling of immunogenomics with protein and spatial information.
Biomarker Identification from Immunogenomics
Immunotherapies are proving very effective in subsets of patients, but unfortunately only a fraction of patients actually respond. This is driving the search for reliable biomarkers for patient selection and personalized therapy.
Anti-CTLA-4 and anti-PD-1 agents have shown distinct predictive biomarker candidates. For example, CD4+ and CD8+ memory T cell subsets play an important role in response to anti-CTLA-4, and are potential biomarker candidates. For anti-PD-1 therapy, NK cell subsets (but not memory T cell subsets) correlate with clinical response to therapy. Improved understanding of the factors needed for effective antitumor response can be used as therapeutic strategies for melanoma and other cancer indications.
The intersection between immunogenomics, cancer genomics, and immunotherapy proves to be very effective in correlating mutational load and response to immunotherapy. For example, patients with melanoma and lung cancer with high mutational load respond to checkpoint blockade therapy. This type of therapy has also been effective in colorectal cancer patients with mismatch-repair-deficiency and high microsatellite instability.
Conflicting reports, however, have undermined the correlation between mutational load and response to immunotherapy. Analysis of melanoma patients has shown that neoantigen burden is not correlated with T cell density, and that indications such as renal cancer carcinoma do respond to immunotherapy despite having a low mutational burden. All these differential responses may explain the existence of intratumor heterogeneity (ITH). Immunogenomic studies are also proving useful in delineating mechanisms of tumor evasion by the immune system, for example, disruption of the interferon γ signaling pathway. Interferon γ can activate other tumor escape mechanisms, such as the upregulation of PD-L1 on the tumor cell surface.
To better understand the interplay between patients' mutations and the immune system, mutant peptides are systematically tested for immunogenicity. This is the ability to activate T cells taken from the same patient. The result of this antigen screening helps in the creation of more personalized immunotherapies, such as tumor-specific vaccines or adoptive T cell therapies. Unbiased genomic screens that use CRISPR-Cas9whole-genome screening also hold great potential for uncovering new resistance mechanisms.
Neoantigens
Antigens that arise in tumor cells due to mutations (neoantigens) allow the immune system to recognize tumor cells as non-self and are therefore a critical factor in triggering a tumor-specific immune response.
The potential and validation of neoantigens is in high demand, and identification is performed by numerous methods. Whole-exome or whole-genome sequencing are used to identify patient-specific non-synonymous mutations. The limitation with these methods is the identification of neoantigens from the sequencing data. This information can, for instance, be used to turn a cold tumor hot, through vaccination, or to educate T cell clones that can eliminate tumors in a patient specific manner. These two approaches are complementary to each other.
Different computational tools have been developed to predict which neoantigens will bind the HLAs expressed on a tumor cell surface with sufficient affinity. However, these technologies are not very accurate and also difficult because of the vast amount of data and very restricted number of validated neoantigens, rather than predicted neoantigens mainly from 0 to 5 which correlates with the mutational load.
Neoantigen based approaches have shown some success. Neoantigen based vaccines induced an antitumor response in rodents, and the clinical potential is now being evaluated. Synthetic RNA-based and peptide-based vaccinations developed via neoantigen are sufficiently immunogenic and significantly benefit melanoma patients. Neoantigens can also be used to engineer specific TCRs for T cells taken from patients ex vivo as adoptive T cell transfer.
High-Throughput T Cell Receptor Sequencing – TCRseq
Adaptive immune cells - T cells and B cells can recognize microbial pathogens (bacteria, viruses, and fungi) and tumors. T and B cells express receptors on their surface, the T cell receptor (TCR) or B cell receptor (BCR), which bind to a specific target and differ from one immune cell to the next. The vast diversity of these receptors allow an individual immune systems to respond against a particular antigen if it presents a threat.
These immune cells not only circulate freely in the blood but infiltrate in tumor tissues, referred to as tissue-infiltrating lymphocytes (TILs). The TIL population differs from the immune cells circulating in the blood, and the two populations of cells and TCRs can be differentiated by high-throughput next-generation sequencing https://science.sciencemag.org/content/352/6291/1337.
Statistics help to compare the TCR patterns between the two different populations across groups of patients being treated for cancer. This can help us assess whether the TCR repertoire inside a tumor differs from that in the circulating blood. These properties may be useful as multi-dimensional biomarkers to monitor tumor progression and therapeutic response.
Future Directions of Immunogenomics in Oncology Drug Development
Immune surveillance and pan-cancer analyses provide information on inter- and intratumoral heterogeneity, which can inform drug response and resistance mechanisms. Another avenue is priming patients' T cells towards tumor neoantigens, based on the accurate selection of targeted neoantigens in combination with immune checkpoint inhibitors that enhance response.
The most suitable therapeutics drugs, which would be based on the selection of specific neoantigens to target and include multiple neoantigens, are yet to be determined. Overall, the identification of novel strategies would be based on the selection of markers which could overcome the therapeutic resistance to immunotherapy by targeting the genetic composition, immune microenvironment, and neoantigens.
Conclusion
Immunogenomics is an emerging field in oncology drug development, deeply invested in answering the key questions related to immunotherapy response and uncovering the key to unlocking resistance. Immunogenomics analysis is already helping to uncover elements such as high mutational load and neoantigens, which have been shown to enhance treatment response.