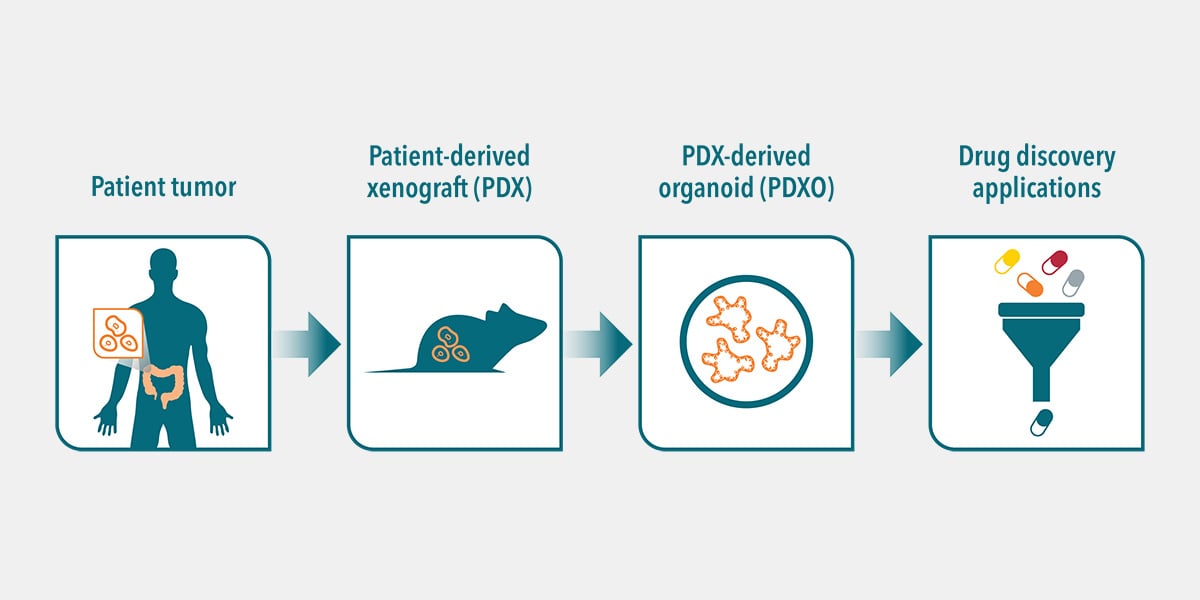
High-content analysis (HCA) is a powerful approach for drug discovery and drug validation studies, especially when combined with highly clinically relevant 3D in vitro organoids. Combining these technologies allows you to develop comprehensive cellular profiles of how your compounds are affecting cellular systems and spatial biology.
As some researchers may be cautious in employing HCA due to the complexities of the technology and downstream analyses, in this post we provide a brief overview of HCA and how we’re using it (in combination with 3D organoids) to support a variety of important projects.
What is High-Content Analysis?
HCA refers to the application of algorithms to high-content imaging (HCI) data. HCI serves as the foundation of HCA by combining automated microscopy with fluorescent detection, which is used for the multiplex visualization and analysis of cellular processes. When used with 2D or 3D in vitro models, HCI can be used to identify treatment effects on the target site, as well as to gain insights in off-target effects in the same assay.
HCI is also readily scalable for high-content screening (HCS) with 3D in vitro organoids (see here for more information on organoids). The combination of HCS and organoids creates a highly predictive empirical platform that offers the best opportunity that the data will have high translatability. With progress in the development of HCA algorithms tailored specifically for HCI and HCS data, a greater number of features and endpoints can be evaluated in complex cellular systems.
A recent study highlights the utility of using organoid biobanks and HCI and HCA in drug discovery. More than 500 therapeutic dual-targeting bispecific antibodies were functionally evaluated by HCI to better understand the complexity of patient-derived organoid (PDO) responses. This drug discovery strategy resulted in the identification of a novel bispecific antibody for colorectal cancer.
Capturing HCA Data
Once an appropriate in vitro model has been selected, organoids are thawed and mechanically sheared to seed organoid fragments in a 384-well plate. Automation is crucial to generate, track, and analyze HCI plates. Small to large scale liquid handling systems (e.g., Apricot S3, Felix, Tecan Fluent) and robotics (e.g., PlateButler®) move stacks of plates in and out of incubators and automated HCI instruments. By making use of ready to go, cryopreserved organoids in combination with liquid handling robotics to seed and expose the organoids to compounds, many plates can be produced per day, as seen below.
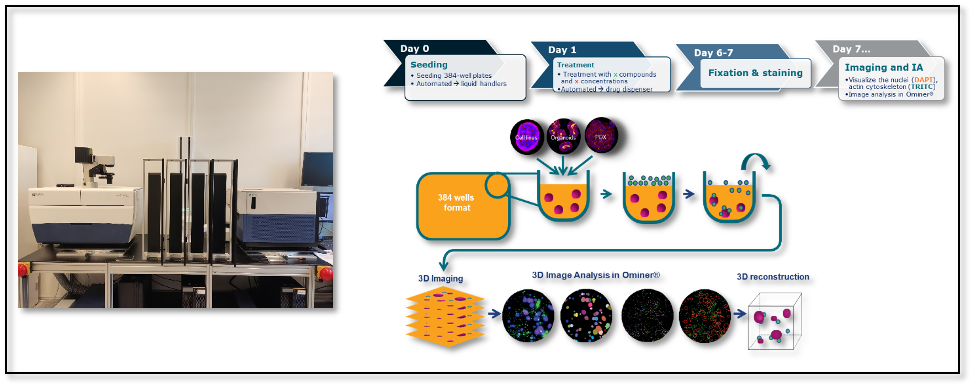
High-content imaging and analysis workflow.
In terms of instrumentation, HCI capture can be carried out using wide-field or confocal microscopes (e.g., Molecular Devices ImageXpress Micro XLS, ImageXpress Micro Confocal) with confocal providing better image and spatial resolution, but at increased cost. A 4x objective is well-suited for standard assays (whole well) while a 20x objective is needed for binding/internalization and MoA assays.
Since organoids are complex 3D structures (ranging between 100-500μm in diameter), z-stack imaging is necessary to obtain an accurate representation of the specimen. This technique captures multiple ‘slices’ through the structures which are then analyzed per stack to capture the in-focus information of the 3D structures.
Depending on the number and size of organoids, fluorescent channels needed (typically up to 4), and total number of plates imaged, massive amounts of data are generated and so storage solutions must also be considered when conducting HCA experiments.
HCA Readouts and Results
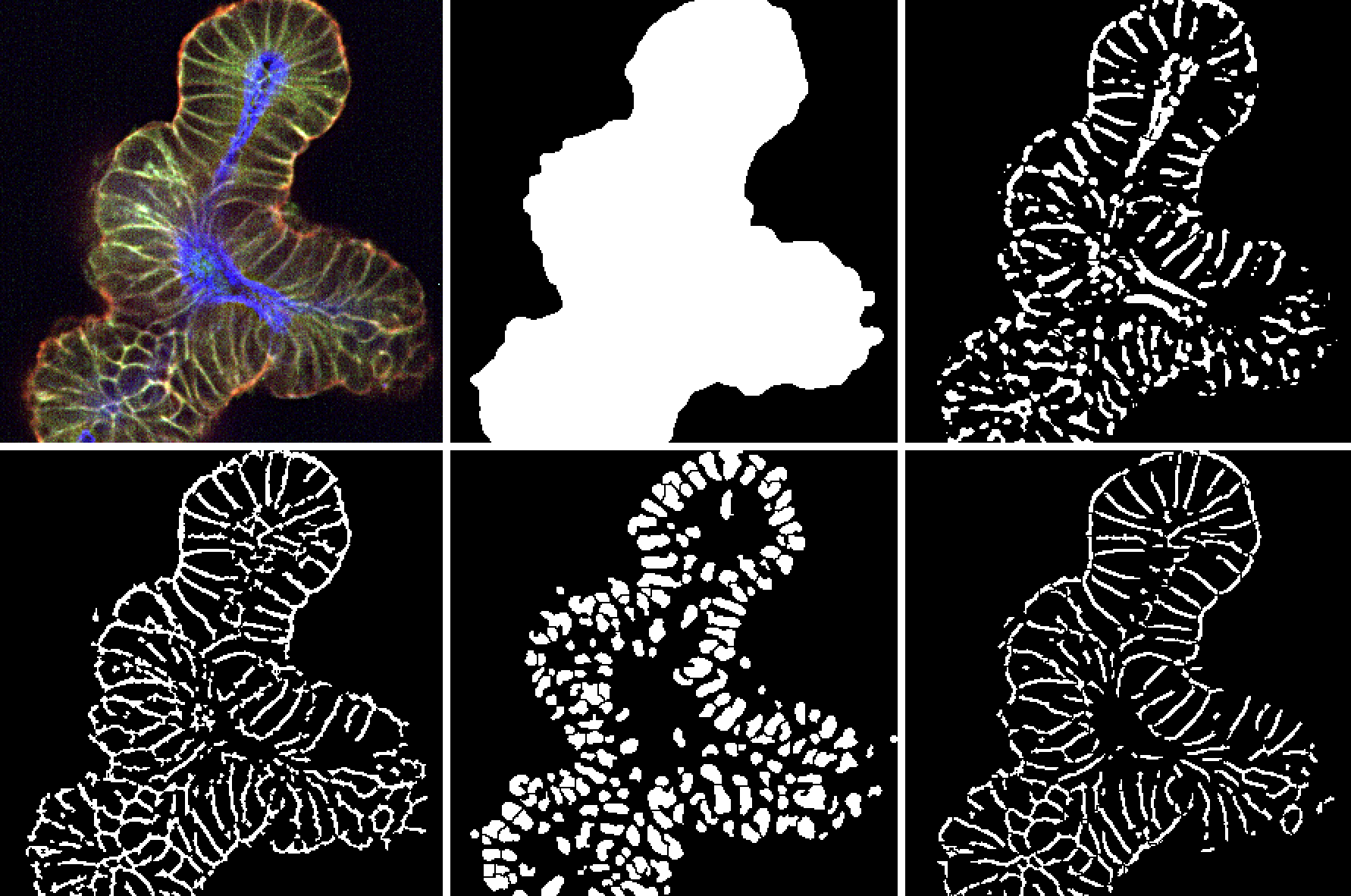
Image quality is critical to the success of HCA and to ensure that the data is reliable. HCA is carried out with a proprietary 3D platform for image analysis and visualization. Initial readouts for a standard assay include raw intensity heatmaps from each fluorescent channel (see below), followed by a thorough quality check for image artifacts that can negatively impact downstream analysis and interpretation.
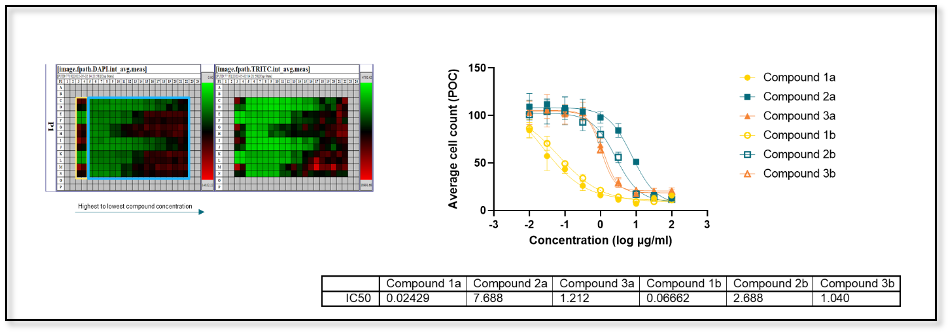
High-content imaging processing and analysis of a standard assay using a proprietary 3D platform. Data analyzed after the image analysis process confirms the preliminary findings from the heatmaps.
HCA significantly expands the amount of information obtained from cellular assays as compared to fixed endpoint assays such as Cell-Titre Glow (CTG) or nuclei count.
For example, nuclei count with HCA includes 300+ data points per model per well, which provides substantial additional biological information that can provide deep insights into the mechanism of a specific compound. Phenotypic and morphological measurements that can be obtained using HCA include:
- Organoid & nucleus: count, size, volume, shape
- Epithelium formation and thickness
- Invasion properties
- Necrosis and apoptosis markers
- Polarity
- Swelling
- Cell cycle
- Internalization
- Therapy/target localization
- IC50, area under-the-curve (AUC)
These readouts can be collected from standard assays testing a small number of compounds in single or broad selection of organoid models. Large scale screening with 1,000 compounds at two different concentrations across a selection of models is also possible.
Analyzing the Data
HCA greatly reduces the time burden and biases associated with manual image analysis, but it does require that accurate and validated algorithms are implemented in the workflow. In addition, while largely automated, HCA does typically require some manual review to ensure the algorithms are performing as expected.
The following are the key steps in HCA:
- Image thresholding to generate an image ‘mask’ that distinguishes signal from background
- Segmentation to define separate objects/organoids, cellular boundaries, and subcellular structures (e.g., nuclei)
- Feature extraction (e.g., fluorescence intensity, lumen width, organoid size, etc.)
- Treatment-level data analysis
Organoid models can have different morphologies and so it is critical that algorithms are capable of accurately capturing this diversity. This can be accomplished by leveraging flexible tools where specific image analysis workflows are available per morphology, as shown in the example below.
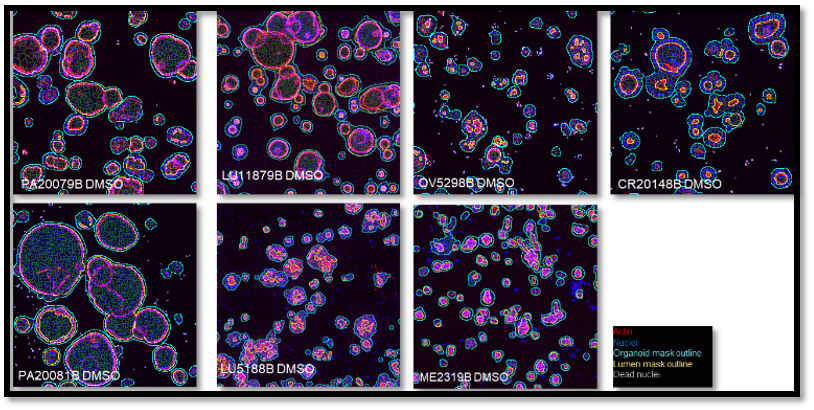
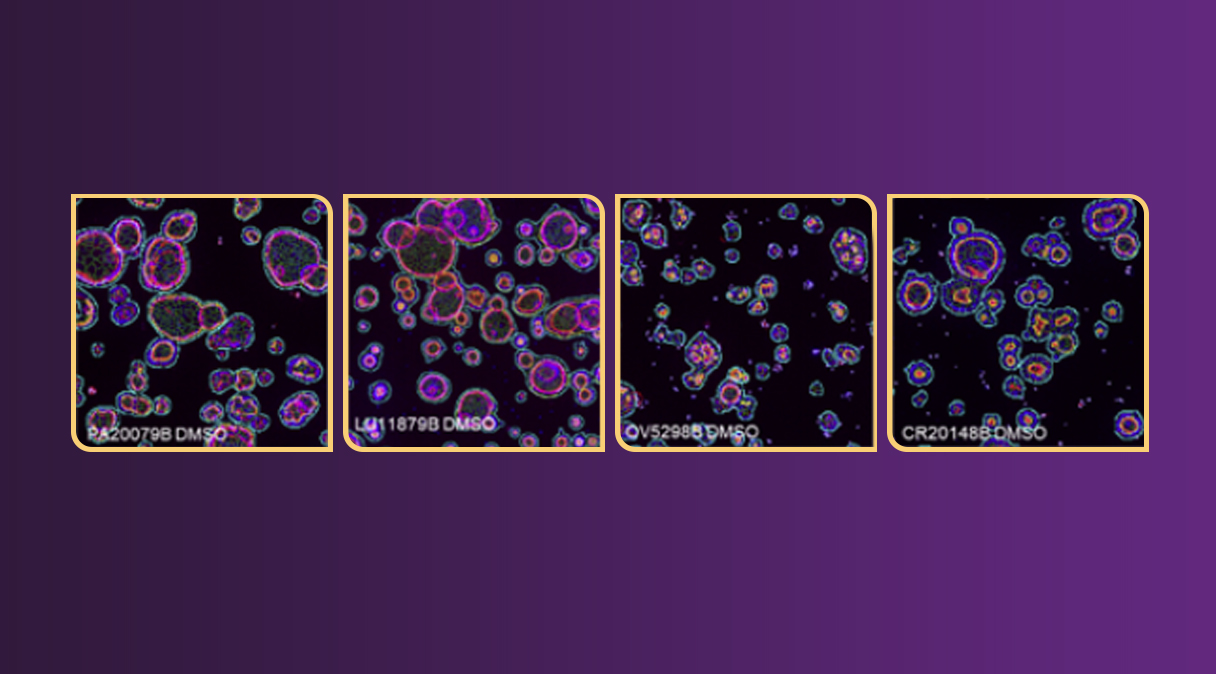
Representative 3D Images of organoids derived from different tumor types (pancreatic, lung ovarian, colorectal, melanoma).
Although organoids derived from human or mouse tumors currently make up the majority of HCA studies, it is important to note that other cell types are amenable to 3D culture.
For example, 3D cultures of myeloid cells have been shown to recapitulate the morphological characteristics observed in vivo. Image analysis with our in-house-developed software successfully identified the most distinguishing phenotypic characteristics for different myeloid cell populations.
Subsequently, a classifier (i.e., an algorithm that automatically categorizes data into one or more classes) was trained with the selected features (e.g., shape, size, and “spikiness”) to create accurate phenotypic profiles for each cell population that can be used to evaluate cellular polarization.
3D culture of human or mouse polycystic kidney cells has also become the standard to model polycystic kidney disease (PKD), a disorder in which clusters of cysts develop in the kidney, leading to loss of function. Cyst formation can be reliably recapitulated in 3D and HCA has successfully been used to measure and understand the nature of swelling in an in vitro setting.
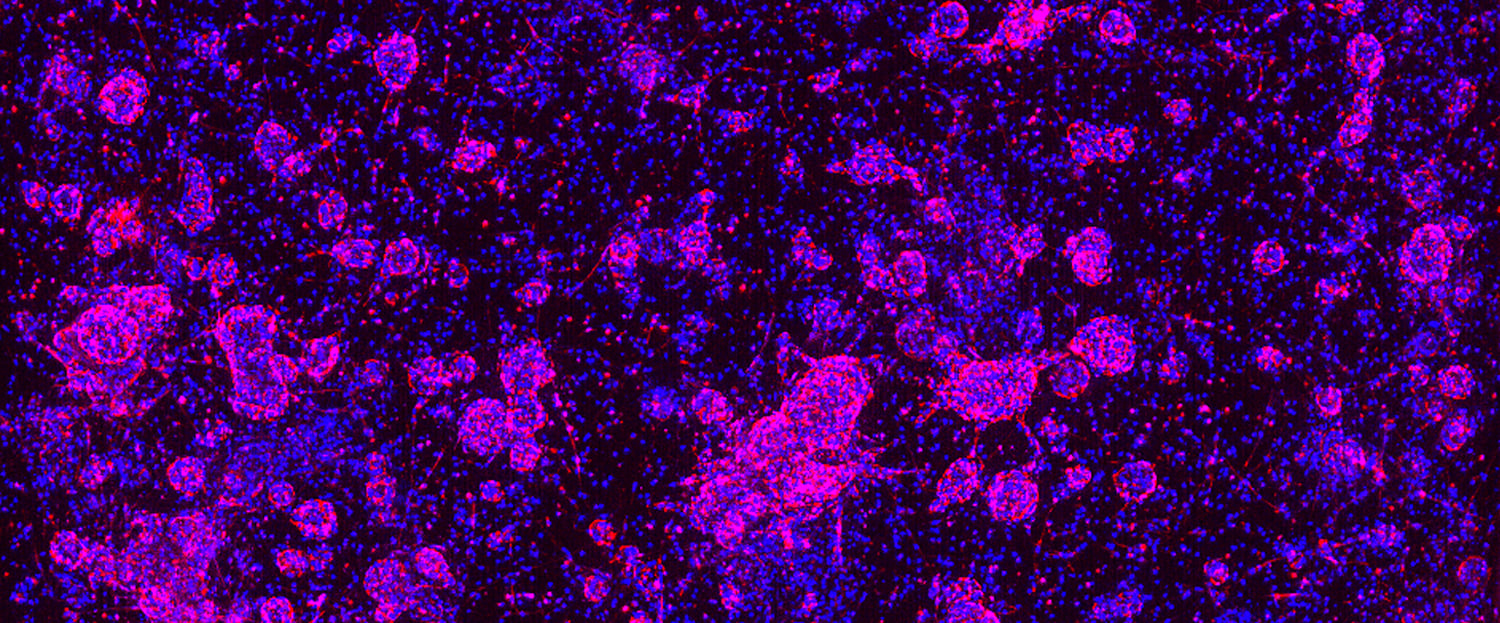
HCA can also capture additional morphological features, such as membrane-bound versus intracellular, in a single assay and up to two markers can be combined in FITC and Cy5 channels. This approach has been used effectively to quantify marker expression or antibody localization and uptake (see example below).
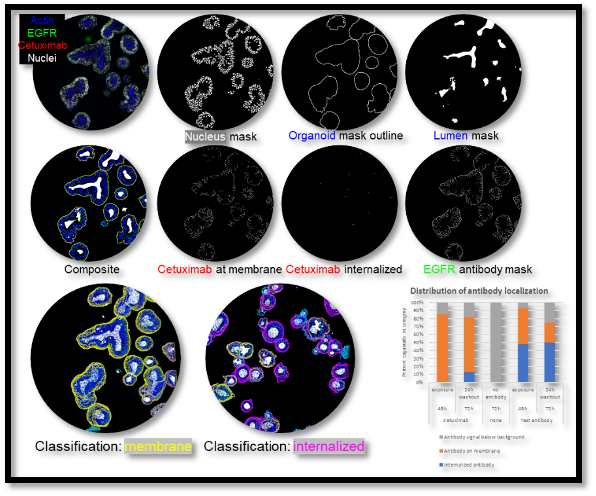
HCA with four fluorescent markers to quantify antibody localization and uptake in organoids.
HCA is not limited to monoculture organoid studies. Complex 3D co-cultures are now commonly being used to evaluate immunotherapy efficacy in the context of an experimental system that contains immune cells, tumor cells, and the microenvironment.
Conclusion
Over the past few years, HCI and HCA technologies have matured greatly, and they are now being used to support drug discovery and drug validation efforts. By combining HCI and HCA with clinically relevant 3D in vitro organoids in large screening studies, researchers can develop more complete cellular profiles in the context of how compounds affect complex cellular systems and spatial biology.
Combining these technologies requires the expertise of highly skilled scientists that understand how to deploy the right tools. If you have questions on how your research program may benefit from a combination of HCI and organoids, please contact us to speak with one of our scientists.