Despite significant progress made over the last 20 years, colorectal cancer (CRC) is still a leading cause of cancer-related death worldwide. Improved preclinical models that will help precision medicine become a reality for patients with CRC and identify biomarkers of response are urgently needed. A team of researcher at the University of Wisconsin Carbone Cancer Center developed new preclinical models for CRC that will help to identify the patients population that will most likely benefit from new therapies.
With recent advances in our ability to genomically profile each patient’s tumors, it has become evident that CRC is as a collection of multiple tumor subtypes, characterized by unique mutation profiles and requiring possibly different treatment approaches.
Each histological type of CRC can harbor two to six different driver mutations. The most frequently mutates genes in CRC are APC, found in 80% of colon cancer cases, TP53 (50%), KRAS (35%-45%), PIK3CA (20%-30%), and BRAF (10%) and their mutation correlates with tumor initiation, progression, and metastasis. In the clinic the mutational status of KRAS, NRAS, and BRAF is frequently assessed to predict whether patients will respond to the anti-epidermal growth factor receptor (EGFR) antibodies cetuximab and panitumumab, as the presence of an aberration in these genes correlates with drug resistance.
Targeting subtypes of CRC, taking a precision medicine approach has proven beneficial for patients whose tumors are BRAF mutant, HER2 expressing, or demonstrate microsatellite instability (MSI). A Phase I study in patients with BRAF mutant CRC has shown that the combination of vemurafenib, cetuximab, and irinotecan results in a 35% response rate, while the combination of trastuzumab and lapatinib benefitted patients with HER2-overexpressing CRC. In addition, pembrolizumab demonstrated a 40% response rate for patients with CRC with high MSI. These early results will be investigated further in upcoming clinical trials.
Preclinical Models of CRC
To make precision medicine a reality for all patients with CRC, we need to better understand the biology behind the molecular profile of each subtype and select the most appropriate therapy accordingly. Preclinical models of CRC have been essential in answering basic questions of tumor biology and, more recently, to investigate the response to anticancer agents. The most common murine models of CRC are the ApcMin mice that nevertheless represent only one particular type of CRC, limiting their predictive strength.
The laboratory headed by Dr. Deming at the University of Wisconsin Carbone Cancer Centre has developed an innovative mouse model of CRC that allows for multiple combinations of mutations to be expressed within a single colon cancer. Using this model, the team can control when the cancer forms, in what tissue, and decide the driver mutation. These tumors can then be followed pre- and post-treatment for biomarker identification.
Using this model, Deming’s team demonstrated the benefit for targeting the PI3K pathway in colon cancers with PIK3CA mutations. This therapeutic approach is now being evaluated in new clinical trials, proving the value of preclinical models for developing targeted strategies that exploit the cancer's mutation profile.
Spheroid Cultures
Genetically modified mouse models (GEMM) are powerful tools to investigate tumor biology and response to therapy, however they are not the only available resource. Three dimensional cell cultures (spheroids) can represent a cost effective alternative to GEMM. Cancer cells grown in 3D form hollow spheres, develop folding, and even crypt-like structures, similar to colon tissue in vivo. The Wisconsin team developed multiple lines of spheroids derived from murine and human colon cancers with defined mutation profiles in which to test new treatment options.
At Crown we have a clear understanding that cancer is not a single disease and that the future of cancer treatments lies in the development of personalized medicine.
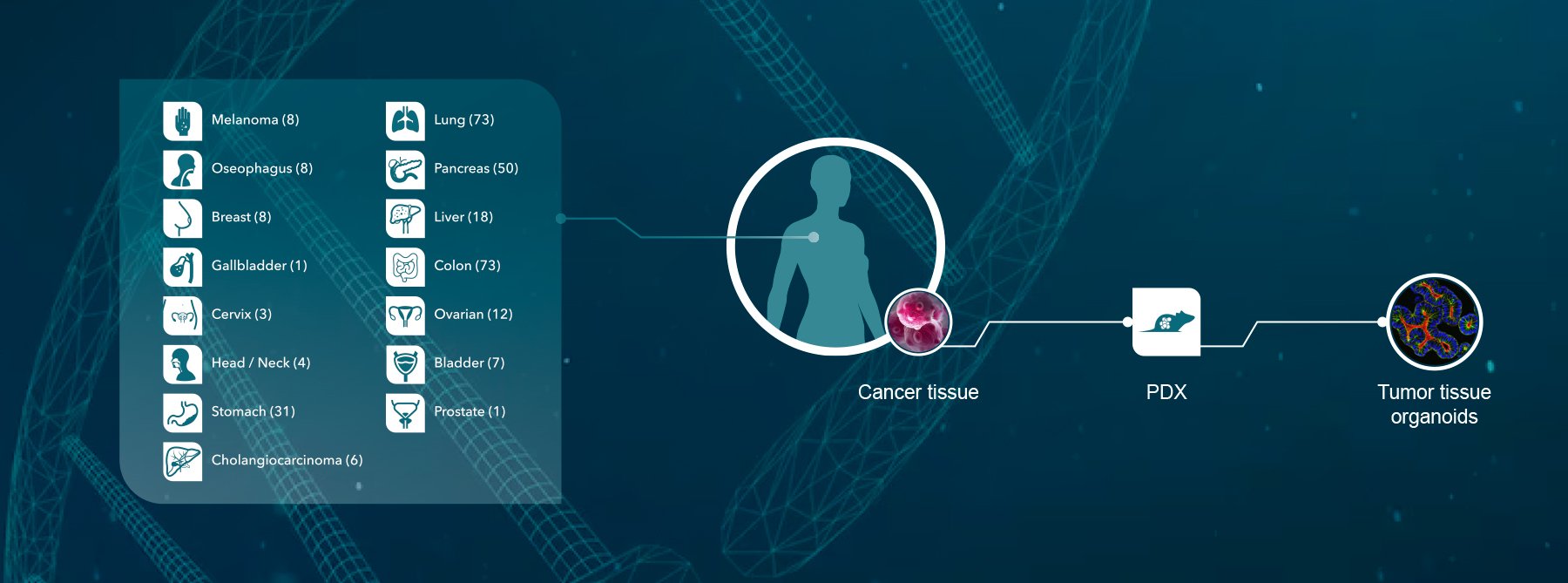
Crown has developed an industry-leading, unique collection of cancer models (including the PDX HuPrime® collection as well as GEMM models), assays, and annotated databases (HuBase™) that can be used by our clients to select clinical candidates by identifying biomarkers of response and to identify genetic signatures (HuSignature™). Furthermore Crown Bioscience has developed a 3D cell culture platform from freshly isolated tumor cells derived from our in vivo models providing a cost effective alternative to in vivo studies.
Contact us today at busdev@crownbio.com for any further questions or information on our in vivo models and 3D cell culture services.